# provide some standard names between DPIRD and SILO for easy merging where
# data values are shared
# not all columns are renamed, but all are listed for clarity
data.table::setnames(
response_data,
old = c(
"station_code",
"station_name",
"longitude",
"latitude",
"elev_m",
"date",
"year",
"month",
"day",
"extracted",
"daily_rain",
"daily_rain_source",
"et_morton_actual",
"et_morton_actual_source",
"et_morton_potential",
"et_morton_potential_source",
"et_morton_wet",
"et_morton_wet_source",
"et_short_crop",
"et_short_crop_source",
"et_tall_crop",
"et_tall_crop_source",
"evap_comb",
"evap_comb_source",
"evap_morton_lake",
"evap_morton_lake_source",
"evap_pan",
"evap_pan_source",
"evap_syn",
"evap_syn_source",
"max_temp",
"max_temp_source",
"min_temp",
"min_temp_source",
"mslp",
"mslp_source",
"radiation",
"radiation_source",
"rh_tmax",
"rh_tmax_source",
"rh_tmin",
"rh_tmin_source",
"vp",
"vp_deficit",
"vp_deficit_source",
"vp_source"
),
new = c(
"station_code",
"station_name",
"longitude",
"latitude",
"elev_m",
"date",
"year",
"month",
"day",
"extracted",
"rainfall",
"rainfall_source",
"et_morton_actual",
"et_morton_actual_source",
"et_morton_potential",
"et_morton_potential_source",
"et_morton_wet",
"et_morton_wet_source",
"et_short_crop",
"et_short_crop_source",
"et_tall_crop",
"et_tall_crop_source",
"evap_comb",
"evap_comb_source",
"evap_morton_lake",
"evap_morton_lake_source",
"evap_pan",
"evap_pan_source",
"evap_syn",
"evap_syn_source",
"air_tmax",
"air_tmax_source",
"air_tmin",
"air_tmin_source",
"mslp",
"mslp_source",
"radiation",
"radiation_source",
"rh_tmax",
"rh_tmax_source",
"rh_tmin",
"rh_tmin_source",
"vp",
"vp_deficit",
"vp_deficit_source",
"vp_source"
),
skip_absent = TRUE
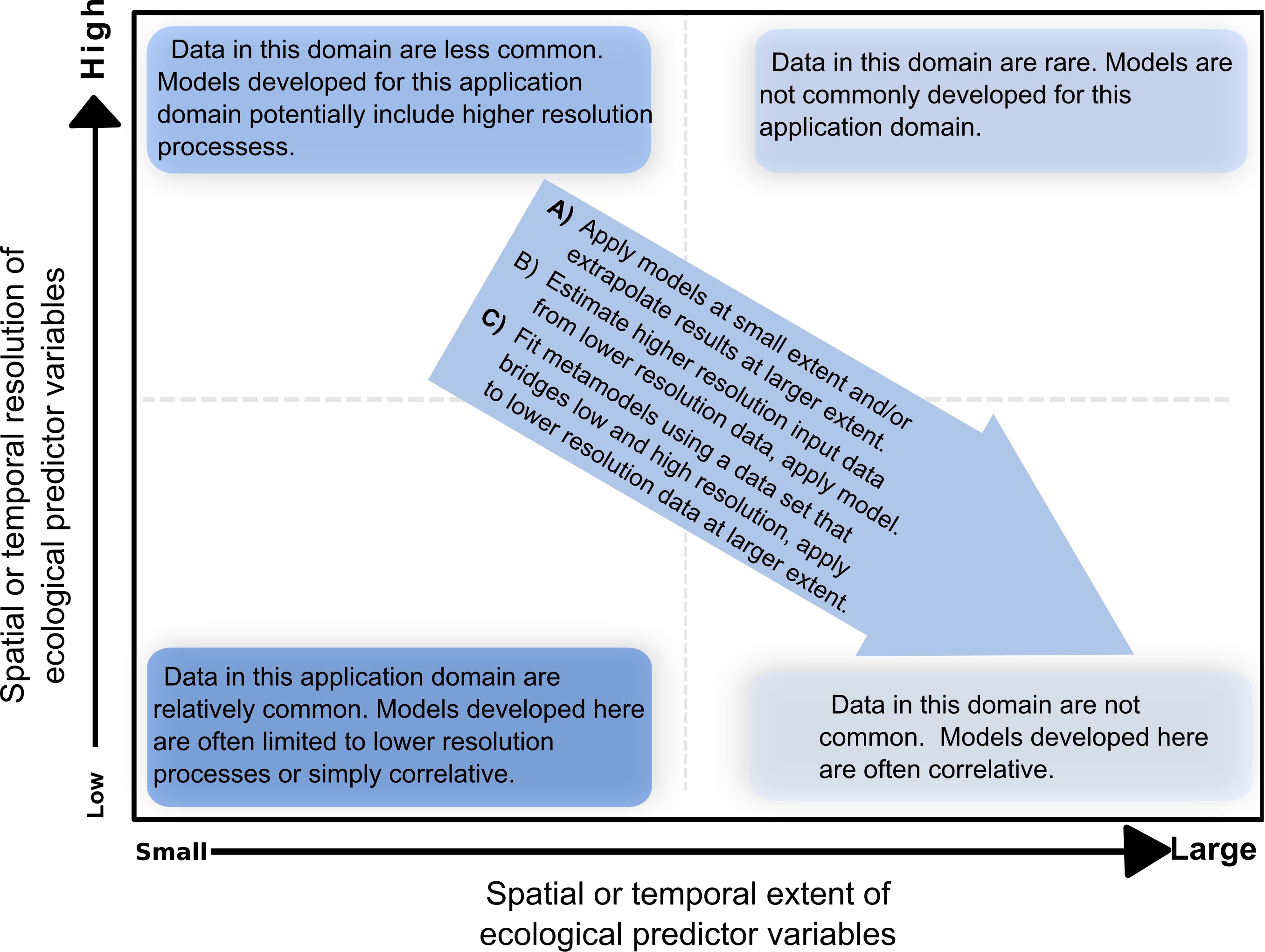
)Climate is what we expect, weather (data) is what we get from APIs

Why Weather Data?

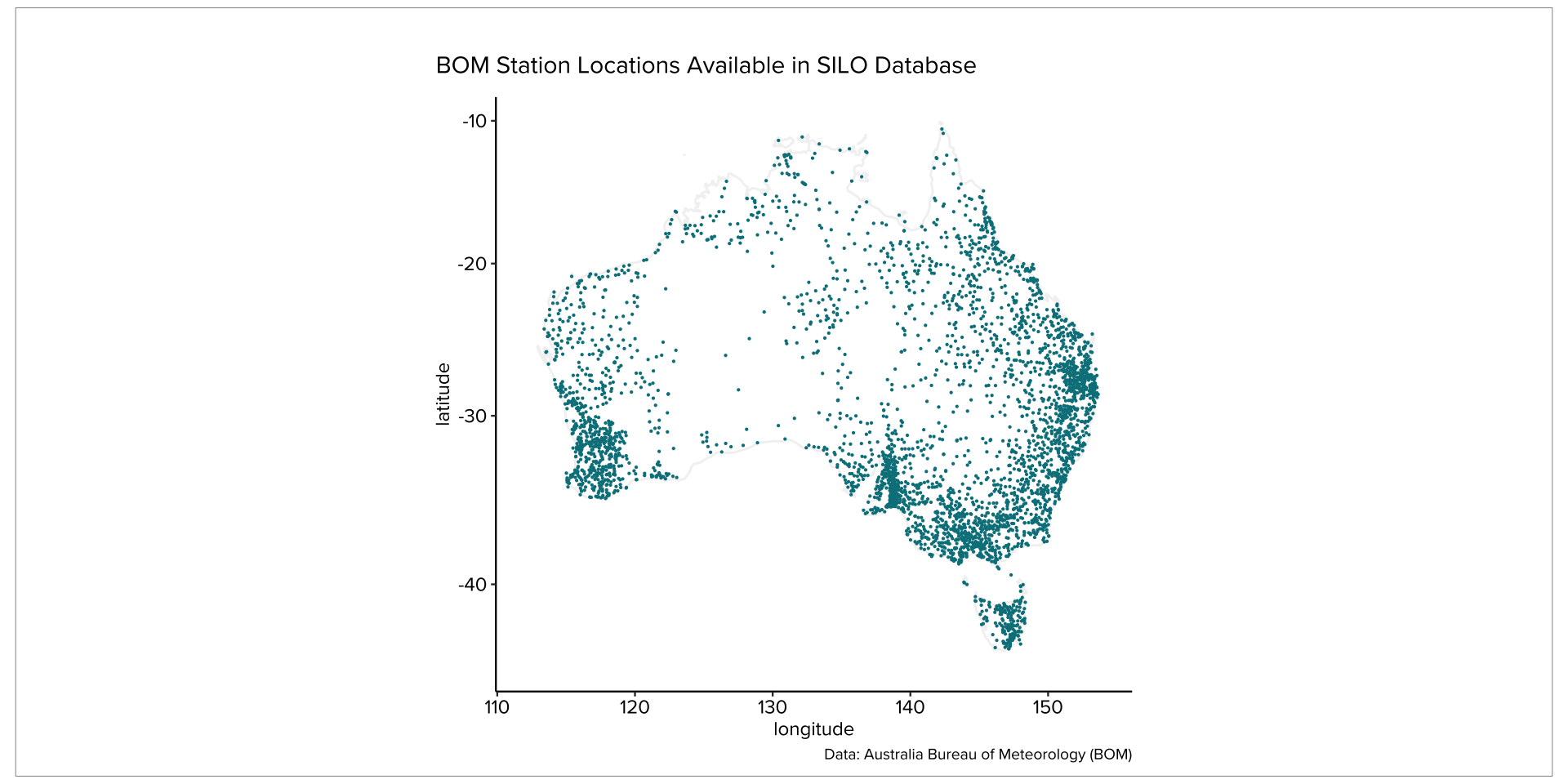
Australian Weather Data
Working With Weather Data
- Sometimes it’s an API (yay!)
- Sometimes it’s HTTP(S) (ok)
- Sometimes it’s (still) FTP (booo)
- NOAA NCEI/NCDC (see also above, HTTP(S))
- BOM (FTP only) – {weatherOz}

Working with APIs
HTTP Client Packages
{crul}  – More developer focused tool
– More developer focused tool
{httr2}  – More interactive use focus
– More interactive use focus
Testing Packages
{vcr}  – Makes testing faster/saves API hits
– Makes testing faster/saves API hits
GovHack 2016

In the Beginning
Error 403 Forbidden
library(bomrang)
get_historical(stationid = "023000", type = "max")
Error in file(con, "r") :
cannot open the connection to 'http://www.bom.gov.au/climate/data/lists_by_element/alphaAUS_136.txt'
In addition: Warning message:
In file(con, "r") :
cannot open URL 'http://www.bom.gov.au/climate/data/lists_by_element/alphaAUS_136.txt':
HTTP status was '403 Forbidden'Full Circle

Screenshot of https://www.longpaddock.qld.gov.au/silo/ 2024.10.09
Welcome {weatherOz}


Lessons Learned
Standardise In-Package
{weatherOz}  Integrates Data From 3 Orgs
Integrates Data From 3 Orgs
SILO
DPIRD
# provide some standard names between DPIRD and SILO for easy merging where
# data values are shared
# not all columns are renamed, but almost all are listed for clarity
data.table::setnames(
out,
old = c(
"station_code",
"station_name",
"year",
"month",
"day",
"date",
"air_temperature_avg",
"air_temperature_max",
"air_temperature_max_time",
"air_temperature_min",
"air_temperature_min_time",
"apparent_air_temperature_avg",
"apparent_air_temperature_max",
"apparent_air_temperature_max_time",
"apparent_air_temperature_min",
"apparent_air_temperature_min_time",
"barometric_pressure",
"battery_min_voltage",
"battery_min_voltage_date_time",
"chill_hours",
"delta_t_avg",
"delta_t_max",
"delta_t_max_time",
"delta_t_min",
"delta_t_min_time",
"dew_point_avg",
"dew_point_max",
"dew_point_max_time",
"dew_point_min",
"dew_point_min_time",
"erosion_condition_minutes",
"erosion_condition_start_time",
"errors",
"evapotranspiration",
"evapotranspiration_short_crop",
"evapotranspiration_tall_crop",
"frost_condition_minutes",
"frost_condition_start_time",
"heat_condition_minutes",
"heat_condition_start_time",
"observations_count",
"observations_percentage",
"pan_evaporation",
"pan_evaporation_12am",
"rainfall",
"relative_humidity_avg",
"relative_humidity_max",
"relative_humidity_max_time",
"relative_humidity_min",
"relative_humidity_min_time",
"richardson_units",
"soil_temperature",
"solar_exposure",
"wet_bulb_avg",
"wet_bulb_max",
"wet_bulb_max_time",
"wet_bulb_min",
"wet_bulb_min_time",
"wind_avg_speed",
"wind_height",
"wind_max_direction_compass_point",
"wind_max_direction_degrees",
"wind_max_speed",
"wind_max_time"
),
new = c(
"station_code",
"station_name",
"year",
"month",
"day",
"date",
"air_tavg",
"air_tmax",
"air_tmax_time",
"air_tmin",
"air_tmin_time",
"apparent_air_tavg",
"apparent_air_tmax",
"apparent_air_tmax_time",
"apparent_air_tmin",
"apparent_air_tmin_time",
"barometric_pressure",
"battery_min_voltage",
"battery_min_voltage_date_time",
"chill_hours",
"delta_tavg",
"delta_tmax",
"delta_tmax_time",
"delta_tmin",
"delta_tmin_time",
"dew_point_avg",
"dew_point_max",
"dew_point_max_time",
"dew_point_min",
"dew_point_min_time",
"erosion_condition_minutes",
"erosion_condition_start_time",
"errors",
"et",
"et_short_crop",
"et_tall_crop",
"frost_condition_minutes",
"frost_condition_start_time",
"heat_condition_minutes",
"heat_condition_start_time",
"observations_count",
"observations_percentage",
"pan_evaporation",
"pan_evaporation_12am",
"rainfall",
"rh_avg",
"rh_tmax",
"rh_tmax_time",
"rh_tmin",
"rh_tmin_time",
"richardson_units",
"soil_temperature",
"radiation",
"wet_bulb_avg",
"wet_bulb_tmax",
"wet_bulb_tmax_time",
"wet_bulb_tmin",
"wet_bulb_tmin_time",
"wind_avg_speed",
"wind_height",
"wind_max_direction_compass_point",
"wind_max_direction_degrees",
"wind_max_speed",
"wind_max_time"
),
skip_absent = TRUE
)Provide Metadata
Include latitude and longitude in data for DPIRD
An excerpt from weatherOz::get_dpird_summaries() lines
...
metadata_file <- file.path(normalizePath(tempdir(), winslash = "/"),
"dpird_metadata.Rda")
if (!file.exists(metadata_file)) {
saveRDS(
get_stations_metadata(which_api = "dpird", api_key = api_key),
file = metadata_file,
compress = FALSE
)
}
...
out <- merge(
x = out,
y = readRDS(file = metadata_file)[, c(1:2, 5:6)],
by.x = c("station_code", "station_name"),
by.y = c("station_code", "station_name")
)
...Simplify API Key Handling
{weatherOz}  makes the API keys easy
makes the API keys easy
- Automate handling of API keys
- Automatic lookup in R session
- Automatic checks that you didn’t use the example key
- Helpful messages if you provide the wrong key
Automate API keys
get_key <- function(service = c("DPIRD", "SILO")) {
service <- match.arg(service)
if (service == "DPIRD") {
DPIRD_API_KEY <- Sys.getenv("DPIRD_API_KEY")
if (!nzchar(DPIRD_API_KEY)) {
.set_dpird_key()
} else {
return(DPIRD_API_KEY)
}
} else {
SILO_API_KEY <- Sys.getenv("SILO_API_KEY")
if (!nzchar(SILO_API_KEY)) {
.set_silo_key()
} else {
return(SILO_API_KEY)
}
}
}Automate API Keys (DPIRD)
.set_dpird_key <- function() {
if (interactive()) {
utils::browseURL("https://www.agric.wa.gov.au/form/dpird-api-registration")
}
stop(
"You need to set your DPIRD API key.\n",
"After getting your key set it as 'DPIRD_API_KEY' in .Renviron.\n",
"DPIRD_API_KEY='youractualkeynotthisstring'\n",
"For that, use `usethis::edit_r_environ()`"
)
invisible("https://www.agric.wa.gov.au/form/dpird-api-registration")
}Automate API Keys (SILO)
Make Sure It’s the Right Key
SILO (E-mail Address)
.is_valid_email_silo_api_key <- function(.api_key) {
pattern <- "\\<[A-Z0-9._%+-]+@[A-Z0-9.-]+\\.[A-Z]{2,}\\>"
if (grepl(pattern, as.character(.api_key), ignore.case = TRUE)) {
return(invisible(NULL))
} else {
stop("For SILO requests you must use your e-mail address as an API key.
You have not provided a valid email address.",
call. = FALSE)
}
}DPIRD (Random String)
.is_valid_dpird_api_key <- function(.api_key) {
pattern <- "\\<[A-Z0-9._%+-]+@[A-Z0-9.-]+\\.[A-Z]{2,}\\>"
if (grepl(pattern, as.character(.api_key), ignore.case = TRUE)) {
stop("For DPIRD requests you must use your DPIRD provided API key.
You (may) have provided your e-mail address, which is used
for the SILO API instead.",
call. = FALSE)
} else {
return(invisible(NULL))
}
}Use a Standard Example API Key
Check for Example Key Use
.check_not_example_api_key <- function(.api_key) {
if (!is.null(.api_key) && .api_key == "your_api_key") {
stop("You have copied the example code and not provided a proper API key.
An API key may be requested from DPIRD or for SILO you must use your
e-mail address as an API key. See the help for the respective functions
for more.",
call. = FALSE)
}
return(invisible(NULL))
}Illustrations of API Keys In Action
Showing Use of Example Key
Showing Use of get_key()
Handle Difficult API Requests Silently
Parse Difficult XML Files
#' Extract the Values of a Coastal Forecast XML Object
#'
#' @param xml_object coastal forecast XML object
#'
#' @return a `data.table` of the forecast for further refining
#' @keywords internal
#' @author Adam H. Sparks, \email{adamhsparks@@gmail.com}
#' @autoglobal
#' @noRd
.parse_coastal_xml <- function(xml_object) {
# get the actual forecast objects
meta <- xml2::xml_find_all(xml_object, ".//text")
fp <- xml2::xml_find_all(xml_object, ".//forecast-period")
locations_index <- data.table::data.table(
# find all the aacs
aac = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::area") |>
xml2::xml_attr("aac"),
# find the names of towns
dist_name = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::area") |>
xml2::xml_attr("description"),
# find forecast period index
index = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::forecast-period") |>
xml2::xml_attr("index"),
start_time_local = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::forecast-period") |>
xml2::xml_attr("start-time-local"),
end_time_local = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::forecast-period") |>
xml2::xml_attr("start-time-local"),
start_time_utc = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::forecast-period") |>
xml2::xml_attr("start-time-local"),
end_time_utc = xml2::xml_parent(meta) |>
xml2::xml_find_first(".//parent::forecast-period") |>
xml2::xml_attr("start-time-local")
)
vals <- lapply(fp, function(node) {
# find names of all children nodes
childnodes <- node |>
xml2::xml_children() |>
xml2::xml_name()
# find the attr value from all child nodes
names <- node |>
xml2::xml_children() |>
xml2::xml_attr("type")
# create columns names based on either node name or attr value
names <- ifelse(is.na(names), childnodes, names)
# find all values
values <- node |>
xml2::xml_children() |>
xml2::xml_text()
# create data frame and properly label the columns
df <- data.frame(t(values), stringsAsFactors = FALSE)
names(df) <- names
df
})
vals <- data.table::rbindlist(vals, fill = TRUE)
sub_out <- cbind(locations_index, vals)
if ("synoptic_situation" %in% names(sub_out)) {
sub_out[, synoptic_situation := NULL]
}
if ("preamble" %in% names(sub_out)) {
sub_out[, preamble := NULL]
}
if ("warning_summary_footer" %in% names(sub_out)) {
sub_out[, warning_summary_footer := NULL]
}
if ("product_footer" %in% names(sub_out)) {
sub_out[, product_footer := NULL]
}
if ("postamble" %in% names(sub_out)) {
sub_out[, postamble := NULL]
}
return(sub_out)
}Use the SILO API
Use the API Interface
...
.query_silo_api <- function(.station_code = NULL,
.longitude = NULL,
.latitude = NULL,
.start_date = NULL,
.end_date = NULL,
.values = NULL,
.format,
.radius = NULL,
.api_key = NULL,
.dataset) {
base_url <- "https://www.longpaddock.qld.gov.au/cgi-bin/silo/"
end_point <- data.table::fcase(
.dataset == "PatchedPoint",
"PatchedPointDataset.php",
.dataset == "DataDrill",
"DataDrillDataset.php"
)
if (.dataset == "PatchedPoint" && .format == "csv") {
silo_query_list <- list(
station = as.integer(.station_code),
start = as.character(.start_date),
finish = as.character(.end_date),
format = .format,
comment = paste(.values, collapse = ""),
username = .api_key,
password = "api_request"
)
} else if (.dataset == "DataDrill" && .format == "csv") {
silo_query_list <- list(
longitude = .longitude,
latitude = .latitude,
start = as.character(.start_date),
finish = as.character(.end_date),
format = .format,
comment = paste(.values, collapse = ""),
username = .api_key,
password = "api_request"
)
} else if (.dataset == "PatchedPoint" && .format == "apsim") {
silo_query_list <- list(
station = as.integer(.station_code),
start = as.character(.start_date),
finish = as.character(.end_date),
format = .format,
username = .api_key
)
} else if (.dataset == "PatchedPoint" && .format == "near") {
silo_query_list <- list(
station = .station_code,
radius = .radius,
format = .format
)
} else {
silo_query_list <- list(
longitude = .longitude,
latitude = .latitude,
start = as.character(.start_date),
finish = as.character(.end_date),
format = .format,
username = .api_key
)
}
client <-
crul::HttpClient$new(url = sprintf("%s%s", base_url, end_point))
response <- client$get(query = silo_query_list)
...However…
# check responses for errors
# check to see if request failed or succeeded
# - a custom approach this time combining status code,
# explanation of the code, and message from the server
if (response$status_code > 201) {
mssg <- response$parse("UTF-8")
x <- response$status_http()
stop("HTTP (", x$status_code, ") - ", x$explanation, "\n", mssg,
call. = FALSE)
}
response$raise_for_status()
# the API won't return proper responses for malformed requests, so, we check
# for the word "Sorry" and parse the response to the user if something slips
# through our user checks.
if (grepl("Sorry", response$parse("UTF8")) ||
grepl("Request Rejected", response$parse("UTF8"))) {
stop(call. = FALSE,
gettext(response$parse("UTF8")),
domain = NA)
}You Don’t Need to Offer Every Option
...
#' @param api_group A `string` used to filter the stations to a predefined
#' group. These need to be supported on the back end. 'all' returns all
#' stations, 'api' returns the default stations in use with the API, 'web'
#' returns the list in use by the weather.agric.wa.gov.au and 'rtd' returns
#' stations with scientifically complete datasets. Available values: 'api',
#' 'all', 'web' and 'rtd'.
...
.build_query <- function(station_code,
start_date_time,
end_date_time,
interval,
values,
api_group = "all",
include_closed,
limit,
api_key)If the Data Isn’t Available…
Make it!
#' Get DPIRD Summary Weather Data in the APSIM Format From the Weather 2.0 API
#'
#' Automates the retrieval and conversion of summary data from the
#' \acronym{DPIRD} Weather 2.0 \acronym{API} to an \acronym{APSIM} .met file
#' formatted weather data object.
#'
#' @param station_code A `character` string or `factor` from
#' [get_stations_metadata()] of the \acronym{BOM} station code for the station
#' of interest.
#' @param start_date A `character` string or `Date` object representing the
#' beginning of the range to query in the format \dQuote{yyyy-mm-dd}
#' (ISO8601). Data returned is inclusive of this date.
#' @param end_date A `character` string or `Date` object representing the end of
#' the range query in the format \dQuote{yyyy-mm-dd} (ISO8601). Data
#' returned is inclusive of this date. Defaults to the current system date.
#' @param api_key A `character` string containing your \acronym{API} key from
#' \acronym{DPIRD}, <https://www.agric.wa.gov.au/web-apis>, for the
#' \acronym{DPIRD} Weather 2.0 \acronym{API}. Defaults to automatically
#' detecting your key from your local .Renviron, .Rprofile or similar.
#' Alternatively, you may directly provide your key as a string here. If
#' nothing is provided, you will be prompted on how to set up your \R session
#' so that it is auto-detected.
#'
#' @section Saving objects:
#' To save \dQuote{met} objects the [apsimx::write_apsim_met()] is reexported.
#' Note that when saving, comments from SILO will be included, but these will
#' not be printed as a part of the resulting `met` object in your \R session.
#'
#' @examples
#' \dontrun{
#' # Get an APSIM format object for Binnu
#' # Note that you need to supply your own API key
#'
#' wd <- get_dpird_apsim(
#' station_code = "BI",
#' start_date = "20220101",
#' end_date = "20221231",
#' api_key = "your_api_key"
#' )
#' }
#'
#'
#' @author Adam H. Sparks, \email{adamhsparks@@gmail.com}
#'
#' @return An \CRANpkg{apsimx} object of class \sQuote{met} with attributes.
#'
#' @family DPIRD
#' @family data fetching
#' @family APSIM
#' @encoding UTF-8
#' @autoglobal
#' @export
get_dpird_apsim <- function(station_code,
start_date,
end_date = Sys.Date(),
api_key = get_key(service = "DPIRD")) {
apsim <- get_dpird_summaries(
station_code = station_code,
start_date = start_date,
end_date = end_date,
interval = "daily",
values = c(
"airTemperatureMax",
"airTemperatureMin",
"panEvaporation",
"rainfall",
"relativeHumidityAvg",
"solarExposure",
"windAvgSpeed"
),
api_key = api_key
)
site <- apsim$station_name[1]
latitude <- apsim$latitude[1]
longitude <- apsim$longitude[1]
apsim[, day := NULL]
apsim[, day := lubridate::yday(apsim$date)]
apsim <-
apsim[, c(
"year",
"day",
"radiation",
"air_tmax",
"air_tmin",
"pan_evaporation",
"rainfall",
"rh_avg",
"wind_avg"
)]
data.table::setnames(
apsim,
old = c(
"radiation",
"air_tmax",
"air_tmin",
"pan_evaporation",
"rainfall",
"rh_avg",
"wind_avg"
),
new = c("radn", "maxt", "mint", "rain", "evap", "rh", "windspeed")
)
apsim <- apsimx::as_apsim_met(
filename = "weather.met.met",
x = apsim,
site = site,
latitude = latitude,
longitude = longitude,
colnames = names(apsim),
units = c("()",
"()",
"(MJ/m2/day)",
"(oC)",
"(oC)",
"(mm)",
"(mm)",
"(%)",
"(m/s)"),
comments = sprintf("!data from DPIRD Weather 2.0 API. retrieved: %s",
Sys.time())
)
return(apsim)
}Examples
PSHB
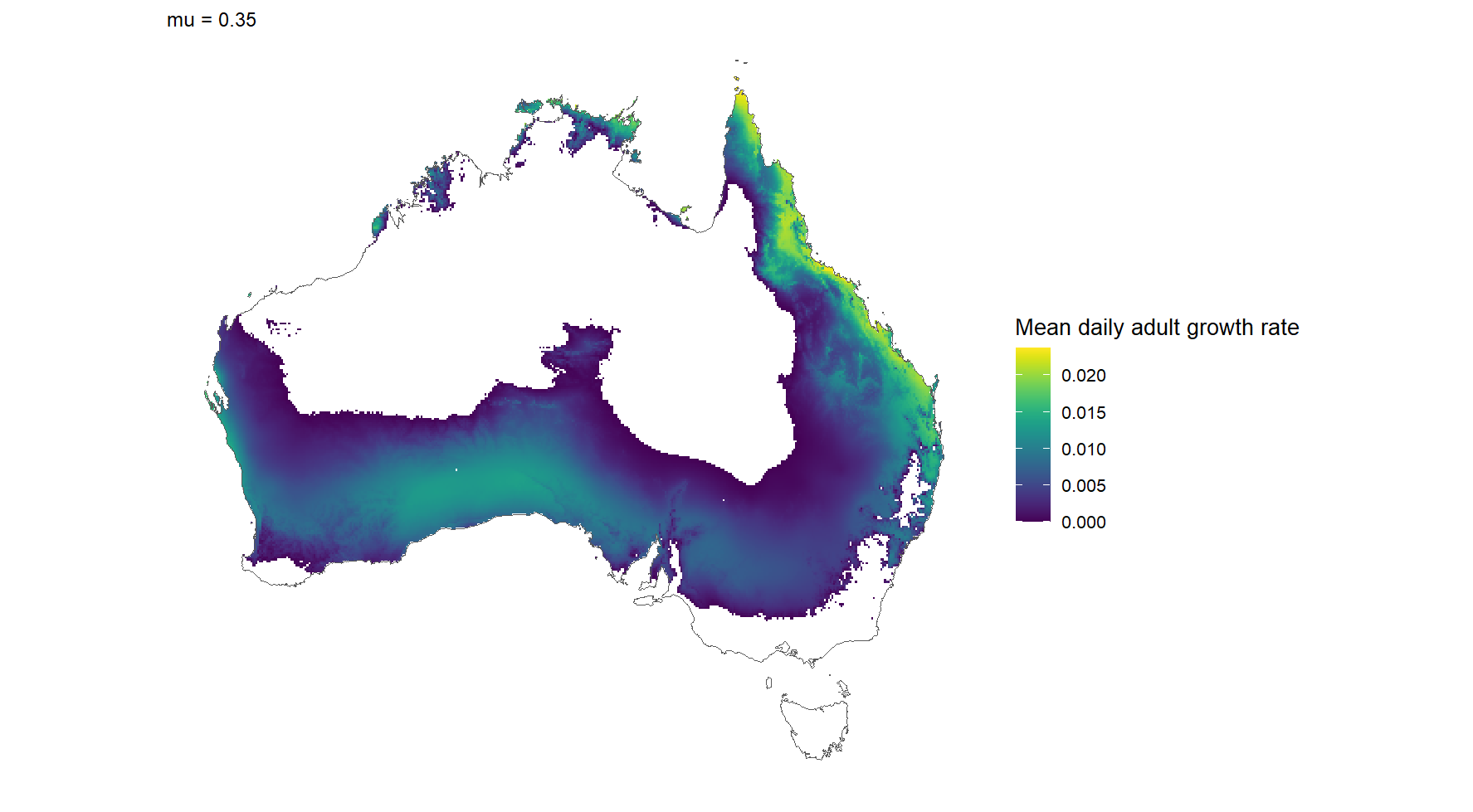
Model output of the posibility of Polyphagous Shothole Borer (PSHB) becoming established. The white areas of Australia indicate that the model suggests that this insect cannot become established. Courtesy Prof. Ben Phillips, Curtin University (unpublished).
Recreating an ABC Article
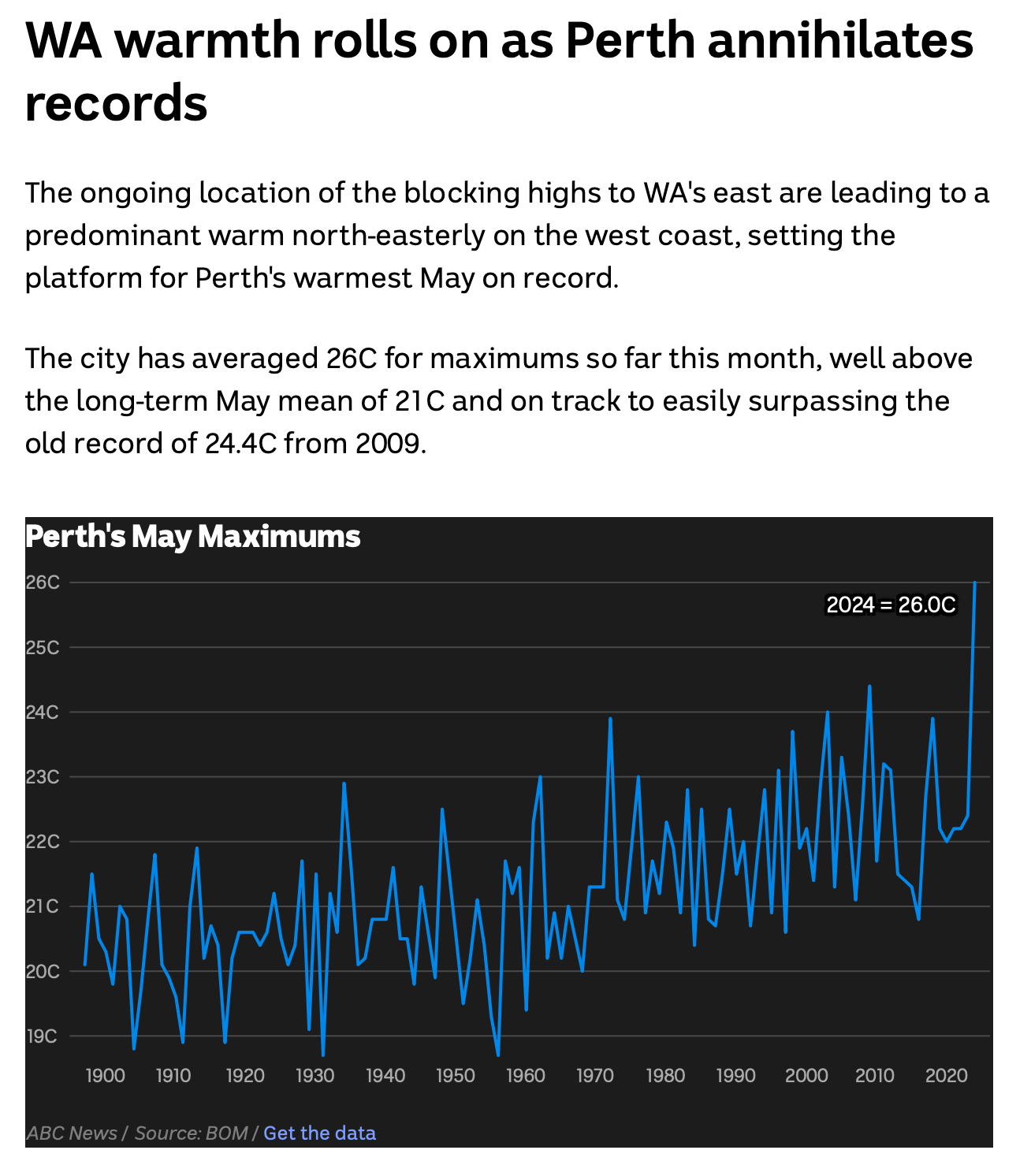
Screenshot from the ABC, https://www.abc.net.au/news/2024-05-18/australia-s-weather-stalls-for-the-second-time-this-month/103862310
Perth Station Locations

Perth Station Data Availability
Perth High Temperatures in May
Full post describing how to build this is available under a CC BY-SA 4.0 Licence. https://adamhsparks.netlify.app/2024/06/02/plotting-perth-month-of-may-high-temperatures-with-weatheroz/
Weather Data
- Useful in many areas of work and research
- Not all sources are created equal, cherish the proper APIs and easy HTTPS access of some
- Make the users’ experience as smooth as possible
- Abstract away as much as possible, e.g., API keys and included meta data
Thank You
Happy Coding!
Slides Available
Codeberg

Rendered Slides, https://adamhsparks.codeberg.page/WOMBAT2024/@main/WOMBAT2024.html#/title-slide
Climate is What We Expect, Weather (Data) is What We Get From APIs by Adam H. Sparks is licensed under CC BY 4.0
